 The main task of this project was to study new approaches and strategies using pattern recognition techniques to improve aspects in the field of bioinformatics and computational biology.
The main task of this project was to study new approaches and strategies using pattern recognition techniques to improve aspects in the field of bioinformatics and computational biology.
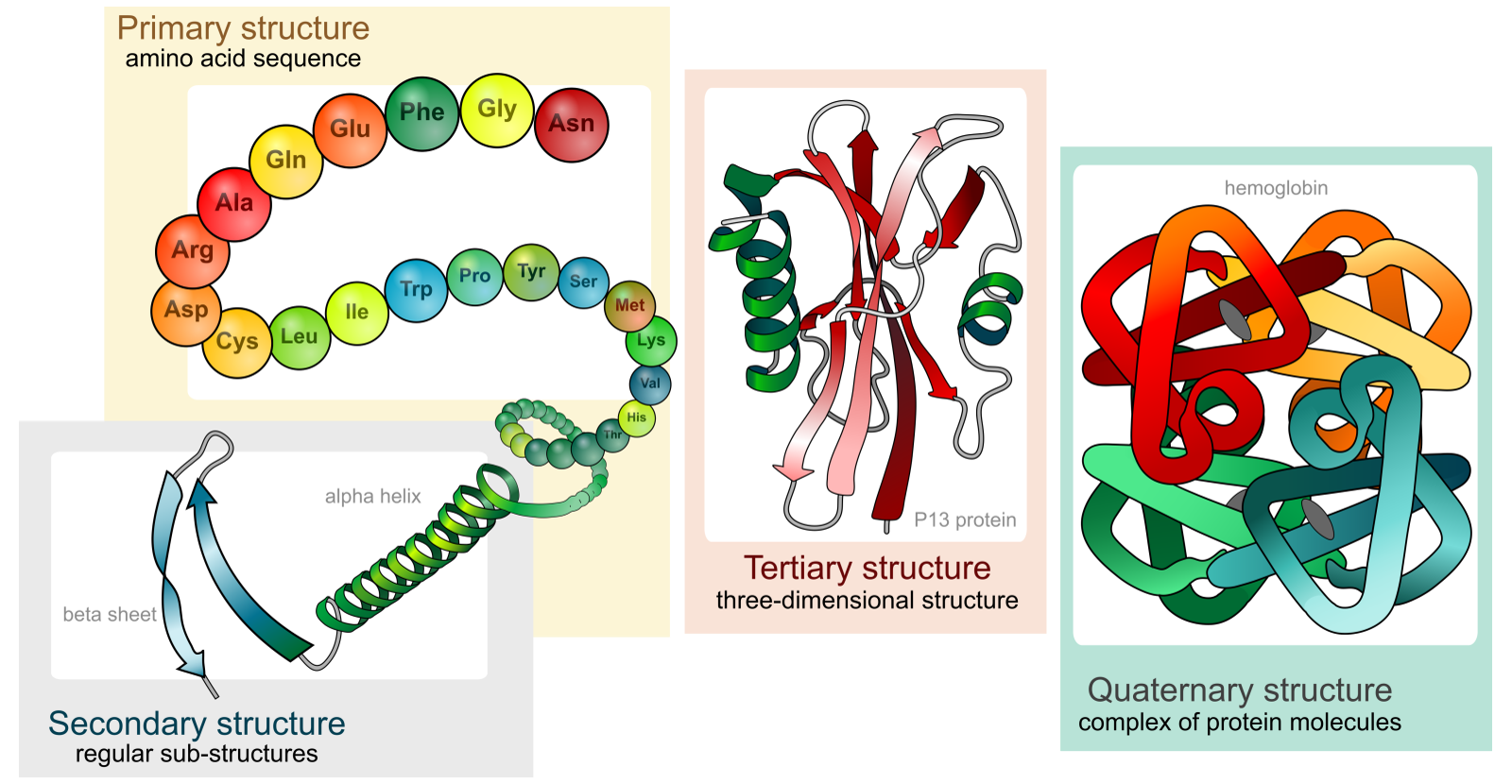
Protein structure define its biological behavior. We worked to identify recurrent motifs, dealing with proteins as a cloud of segments, expecting big data. Our goal was to discover new motifs with an innovative geometrical method.
We applied segmentation algorithms to search areas of interest (active sites) in protein.
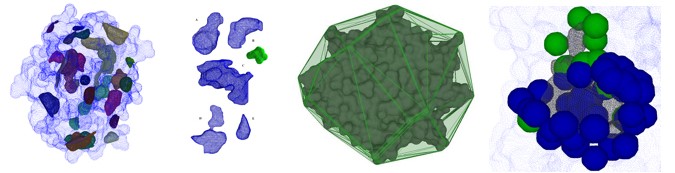
Extended Gaussian image: Histogram of the orientations of a surface represented on a unitary sphere and comparison Pocket - Ligand.
We aligned the models for the docking protein-ligand.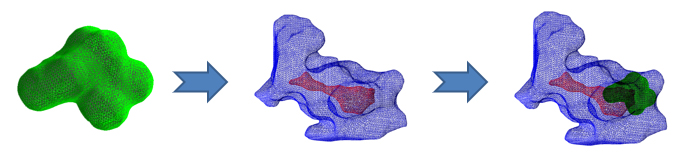
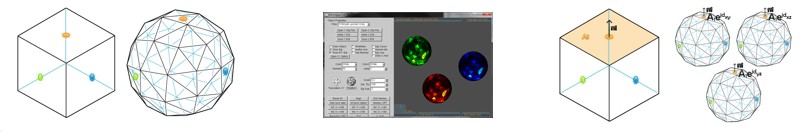
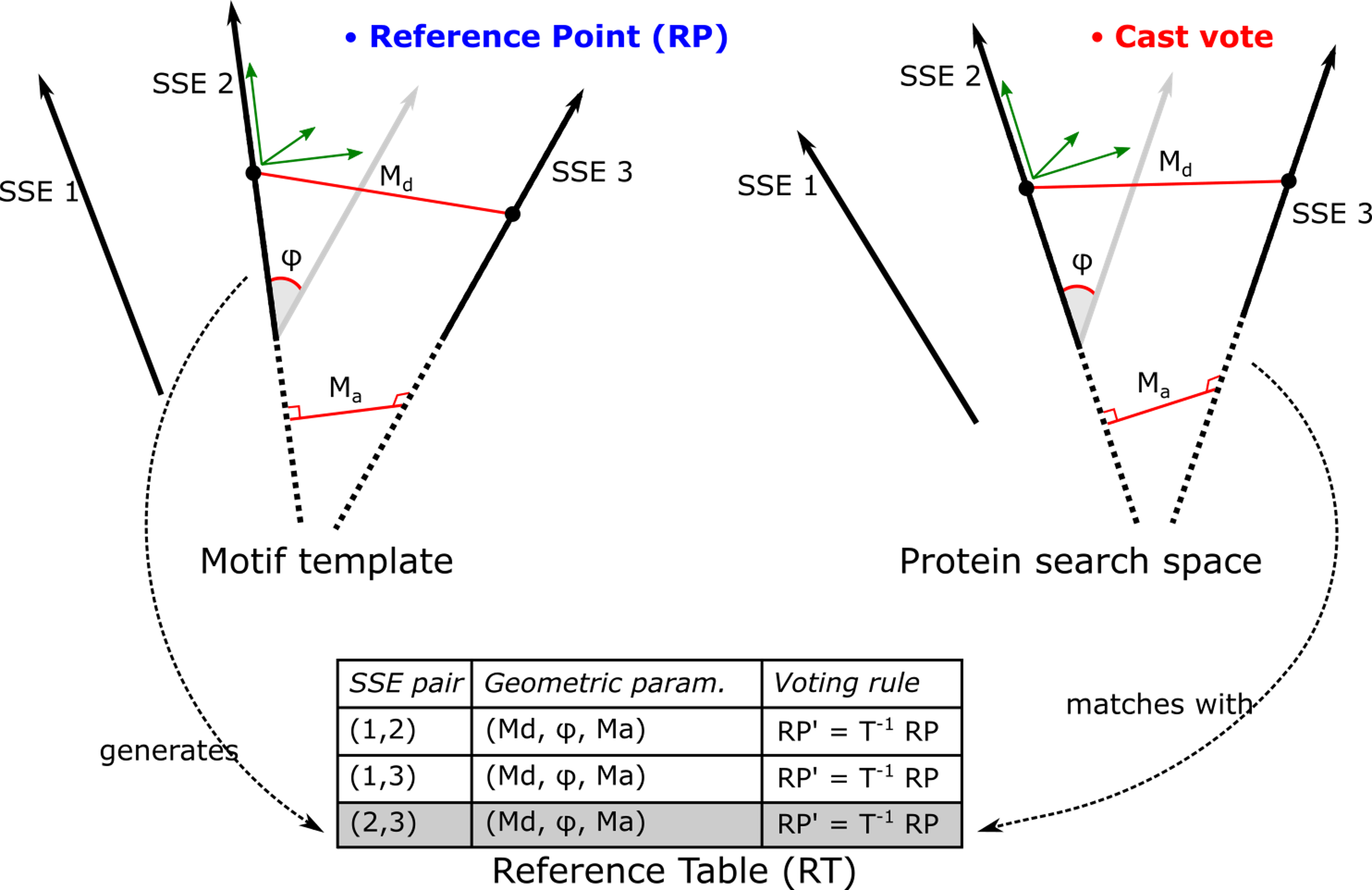
Using Hough Transform we developed the Cross Motif Search algorithm with MP and MPI for parallelism.
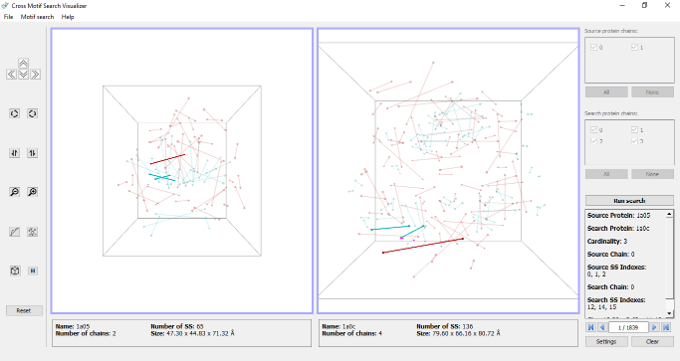
Motif Visualizer is an Open Source OpenGL GUI to improve usability, developed in collaboration and validation with biologists.
 In this context, in 2012, our laboratory has contributed to the TCBionformatics - GIRPR Technical Committee.
In this context, in 2012, our laboratory has contributed to the TCBionformatics - GIRPR Technical Committee.