This research is the focus of the PhD thesis of Gianluca Gerard, developed during the XXXIII cycle of the PhD School in Electronics, Computer Science and Electrical Engineering of the University of Pavia, completed in 2020.
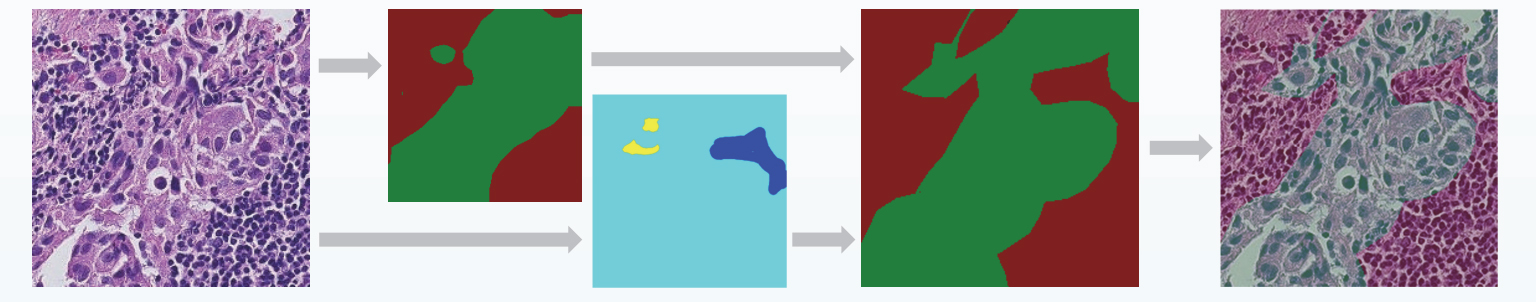
Breast cancer is the most common form of cancer among Italian women. The prognosis depends on whether the cancer has spread to other organs. Sentinel lymph nodes are the target organs primarily reached by metastasizing cancer cells. The diagnosis is conducted by cutting slices from the sentinel lymph nodes, fixing them on glass slides, staining the tissues and, for a digital diagnostic pipeline, digitalizing the slides, to obtain Whole Slide Images (WSIs).
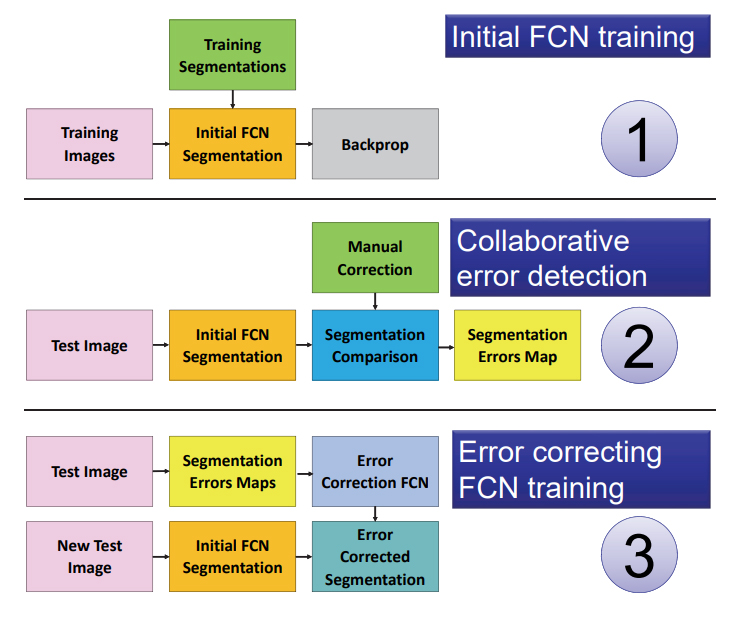
The goal is to design an automated segmentation method to support the pathologists in the diagnosis of WSIs able to improve its performances based on a limited set of error corrections (provided by the pathologist).
We use a ResNet50 Dilated FCN [1] as our main network. The classical ResNet50 architecture is modified such that the last three residual blocks have the standard convolutions replaced by dilated convolutions. The last fully connected layer is replaced by a convolution layer followed by bi-linear up-sampling.
The training images are derived by segmenting WSIs in a regular grid of patches, each 448x448 pixels. Patches with no tissue are discarded by a filter applied to the HSV channels, and the whole network is trained to classify the images with three labels: background, normal tissue and lesion.
Due to the heavy imbalance of normal patches vs patches with lesion we downsample the normal patches and we only process WSIs that contain at least one lesion. Our interactive pipeline then takes inspiration from [2] as we use a separate path to identify discrepancies between the tool’s segmentation (host network) and the manual pathologist segmentation.
The error correction network is based on a recently introduced Fully Convolutional Network (FCN) that learns how to segment new categories with very few input exampexamples [3]. This new FCN is called conditional FCN and it achieves excellent results even if conditioned on single pixels instead of dense segmentations. The error correction network is the same FCN with a conditional path that learns to recognize, segment and reclassify misclassified regions between the pixels where the automatic segmentations agree with the manual segmentation from the ones that doesn’t.
The trained conditional FCN is used on the training set to correct the training labels and to correct its parameters.
We devise a new framework for interactive correction of automated segmentation of lesions in lymph-nodes. Our method relies on a recently introduced enhancement to the Fully Convolutional Network [3] that allows to segment regions of new classes starting from very few samples of the new class. In our framework this allows to pass segmented error regions to the segmentation network and use it as an error detector and classifier. Once the error correction is applied to the original training dataset the network can adapt and change its behavior according to the correction provided by the pathologists.
Our framework is a novel application of co-FCN in the field of biomedical images.